Ontologies are formal and explicitly defined concepts used in specific domains. They are commonly used in biomedical fields to uniform the representation of concepts. For example, in hospitals, the primary care systems encode patient records in controlled vocabularies or ontologies; the dataset researchers get uses ontologies for event registration; and there are ontologists, the people who are behind the work of defining the concepts and their semantic relationships in the ontologies. They are of dominant position in biomedical fields for both practical use and research.
However, the technologies powering the ontologies are not keeping up as good as other fields. There are hundreds of different software, mobile apps, web services or libraries to edit a .doc file, but when we think of ontology files, the first and probably only tool we could think of is Protege. In Python, there is owlready2, and that about sums up the commonly used tool kits. If we want to get ontology information from online services, it’s even more complicated: several gigantic services dominates the ontology service world, such as BioPortal, NHS system, HGNC system, and NIH UMLS. Each provides a collection of different ontologies, uses different API formats, requires a dedicated API key, and is quite complicate to interoperate.
Working in University of Leicester as a researcher in genetics department, part of my job is to deal with ontologies and semantic similarity between ontology concepts. From last year, I decided that instead of coding separate ontology reading/parsing/querying pipelines for different tools, I will make one dedicated service for these types of applications. Not just a tools to fetch ontology concept data, but as a research kit to explore mapping, similarity, graph, and even AI tools empowered by ontologies. From my own experiences I concluded that for a programmer who haven’t had any training in bioinformatics before, to use ontology and work with semantic similarity, the typical troubles are
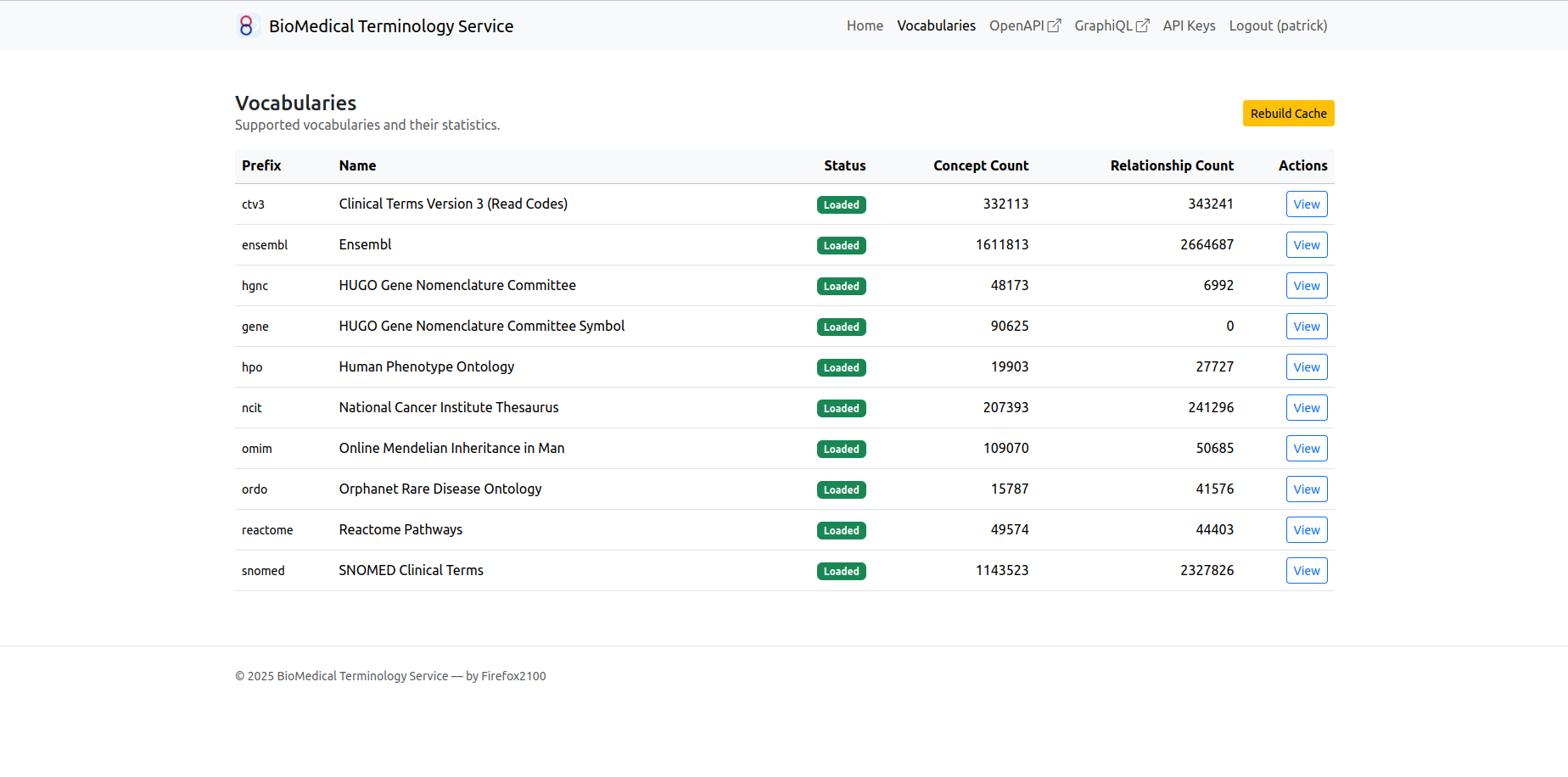
- Not familiar with the release structure of an ontology. HPO is released in OWL format and so is Orphanet data, but SNOMED is released in RF2, OMIM in CSV file, and Reactome in a database dump. If the requirement is to work with one ontology, then it’s manageable; however if a project require its developers to quickly support a large amount of ontologies for simple usage like term auto-completion, it would take them a long time just to figure out how to read the release files.
- Ontologies are graphs, and normally programmers don’t have to deal with something that is explicitly a graph. All operations on an ontology such as finding descendants, getting similar/neighbour terms, cross-ontology mapping, etc. treats it as a graph. This involves using a graph database, designing an API that is capable of retrieve data recursively, and model the relationships accordingly.
- No single source of data. If you want to download HPO, you should head over to their GitHub repository, where they keep the ontology and some mapping files. However, if you also want to work with ORDO, you’d go to Orphanet data platform — only to find that their download procedure is hard to follow and cannot automate. Places like BioPortal offers many ontologies downloading and online querying, but it’s still missing on final step — no direct mapping offered in between them.
- Heavy computation load and fast iterations. Some ontologies are big — they could contain over a million concepts. It’s not uncommon in the development phase to read it, process it, store in database, later just to find out that you missed a property in a filter list and have to do it again, which takes another 30 minutes to finish.
With this in mind and many iterations of codes, I built a software prototype and published it on GitHub.

It’s a web API service that serves the ontology data with a consistent API, plus other advanced features such as:
- Unified semantic similarity scores between concepts calculated with different methods, including well-established ones like Resnik/Lin’s/Relevance method, bleeding-edge ones like Onto2Vec, and some personal experimental ones.
- High-efficiency query support and horizontal scaling. These type of services typically serves a large amount of users, and needs to be queried repetitively. Efficiency becomes an important consideration. This software is designed to maximise query speed, while also allowing horizontal scaling, so that on a cloud environment it can serve hundreds of thousands of queries at the same time, if the infrastructure permits.
- Mapping in between ontologies, even going through intermediate ones. For example, NIH released SNOMED-ORDO mapping, while ORDO has gene symbol alignment mapping, so in theory, a gene symbol can be mapped to SNOMED, if the mapping path exists in both of the releases.
- GraphQL API support for large-scale or on-demand graph traversal. Since the ontologies are graphs, it’s natural to use GraphQL for querying the data.
- Support beyond ontologies. Other vocabularies that are not exactly an ontology, such as Ensembl gene database and Reactome pathway database, can also be loaded and are officially supported. This allows it to handle use cases in various fields.
- Modular design with plugin-style support. All vocabularies, annotation/mapping and semantic similarity algorithms are modular. It’s easy to add another module for another mapping, another vocabulary, or a different method of calculating similarity.

The software is of course still a prototype, and needs many finishing touches for being robust and efficient, or add other community-demanded features. However, I would like to share the existence and development progress of it with everyone, with the hope that it may be useful in even the smallest form. If it’s just to play around with it, find some example code to see how to read from an ontology release file, or actually use it in some other projects, I’d be more than happy if it makes your life a bit easier.
I will continue to work on this project, and please leave a comment or submit an issue if you think it could use some features, or some bug fixes. Stay tuned for the updates and usage examples of the Biomedical Terminology Service!
Leave a Reply